Generate Deeper Insights with Protein Identification on Platinum® Pro
Deeply interrogate individual proteins with high resolution using Next-Gen Protein Sequencing.™
Learn More About Platinum ProWhy choose Next-Gen Protein Sequencing™ (NGPS™) for protein identification?
Complete protein identification prior to additional experiments
Identify unknown bands from your gels
Determine which proteins are in a biological sample like serum
Identify potential contaminants from protein production
Verify that your antibody is successfully isolating the target protein
The ability to accurately identify proteins is essential to exploring function, understanding impact to biological process, and uncovering disease mechanisms.
Next-Gen Protein Sequencing with single-amino acid resolution offers the precision needed to avoid drawing inaccurate conclusions.
When analyzed with NGPS, your protein is digested into peptides and individually distributed across our sequencing chip. Each peptide is sequenced independently and generates distinct kinetic signatures that come from every single amino acid in your sequence.
These signatures are then mapped to a proteome-wide inference panel, enabling you to identify the proteins present in your sample. NGPS enables single-molecule level characterization of individual proteins from biological samples with precision and sensitivity.
Talk With an ExpertExplore our resources to discover how Platinum Pro can enhance your protein identification.
-
Next-Gen Protein Sequencing™ Enables Accurate AAV Serotype Profiling and Mixture Resolution for Gene Therapy — APP NOTE
Adeno-associated viruses (AAVs) are widely used as delivery vectors in clinical gene therapy, with each serotype exhibiting unique capsid protein sequences that influence tissue tropism, transduction efficiency, and immune response. Ensuring the identity and purity of AAV preparations is essential during manufacturing and quality control, particularly when closely related serotypes are involved. In this study, we demonstrate the use of Next-Gen Protein Sequencing™ (NGPS™) to identify AAV8 and AAV9 capsid proteins by detecting three serotype-specific peptides capturing amino acid differences at position 105, 315, 539, and 540. NGPS enabled reliable identification of each serotype and unambiguous detection of each serotype from a mixture, with sensitivity sufficient to detect AAV8 and AAV9 at levels of 10% in a mixture based on distinct kinetic signatures. These results highlight the utility of NGPS as a high-resolution analytical tool for AAV serotype characterization and monitoring of contamination.
-
Identifying Cancer-Specific Proteins Using Next-Gen Protein Sequencing — CASE STUDY
Sanavia Oncology is focused on developing antibody therapeutics targeting novel cancer markers, and plans to utilize Next-Gen Protein Sequencing™ to better characterize novel cancer targets, which is a crucial tool in their target identification workflow.
-
Next-Generation Protein Sequencing and individual ion mass spectrometry enable complementary analysis of IL-6 — PUBLICATION
The vast complexity of the proteome currently overwhelms any single analytical technology in capturing the full spectrum of proteoform diversity. In this study, the team at Northwestern University evaluated the complementarity of two cutting-edge proteomic technologies—single-molecule protein sequencing and individual ion mass spectrometry—for analyzing recombinant human IL-6 (rhIL-6) at the amino acid, peptide, and intact proteoform levels. For single-molecule protein sequencing, they employed the Platinum® instrument. Next-Generation Protein Sequencing™ (NGPS™) on Platinum utilizes cycles of N-terminal amino acid recognizer binding and aminopeptidase cleavage to enable parallelized sequencing of single peptide molecules. They found that NGPS produces single amino acid coverage of multiple key regions of IL-6, including two peptides within helices A and C which harbor residues that reportedly impact IL-6 function. For top-down proteoform evaluation, the team used individual ion mass spectrometry (I2MS), a highly parallelized orbitrap-based charge detection MS platform. Single ion detection of gas-phase fragmentation products (I2MS2) gives significant sequence coverage in key regions in IL-6, including two regions within helices B and D that are involved in IL-6 signaling. Together, these complementary technologies delivered a combined 52% sequence coverage, offering a more complete view of IL-6 structural and functional diversity than either technology alone.
Northwestern University’s Neil Kelleher and his team have a published article, featured in Springer’s Analytical and Bioanalytical Chemistry journal. The paper highlights the synergy of complementary protein detection methods to more comprehensively cover protein segments relevant to biological interactions.
-
Protein Sequencing With Single-Amino Acid Resolution Discerns Peptides That Discriminate Tropomyosin Proteoforms — PUBLICATION
Protein variants of the same gene–proteoforms–can have high molecular similarity yet exhibit different biological functions. Thus, identifying unique peptides that unambiguously map to proteoforms can provide crucial biological insights. In humans, four human tropomyosin (TPM) genes produce similar proteoforms that can be challenging to distinguish with standard proteomics tools.
Dr. Gloria Sheynkman and her team at the University of Virginia School of Medicine have a published article in the Journal of Proteome Research showcasing the capabilities of the Platinum® Next-Gen Protein Sequencer™ in advancing their research in this complex area of study.
-
Comparison of Protein Sequencing Analysis of CDNF, IL6, and FGF2 on Platinum and Mass Spectrometry — APP NOTE
In this application note, we compare protein sequencing analysis of CDNF using Quantum-Si’s Platinum protein sequencing platform with mass spectrometry. The Platinum workflow offers an accessible and convenient solution for protein identification without the need for expensive equipment and expertise allowing researchers to gain deeper proteomic insights and make informed decisions about their protein samples.
Download the application note now to unlock a new level of understanding in your protein research.
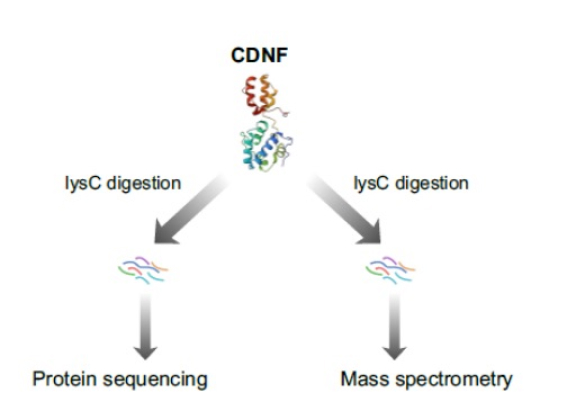
While both mass spectrometry and the Quantum Si platform correctly identified the CDNF protein, the workflow was dramatically different.
Read the application note to see how the Platinum workflow can save you time and effort without compromising results.

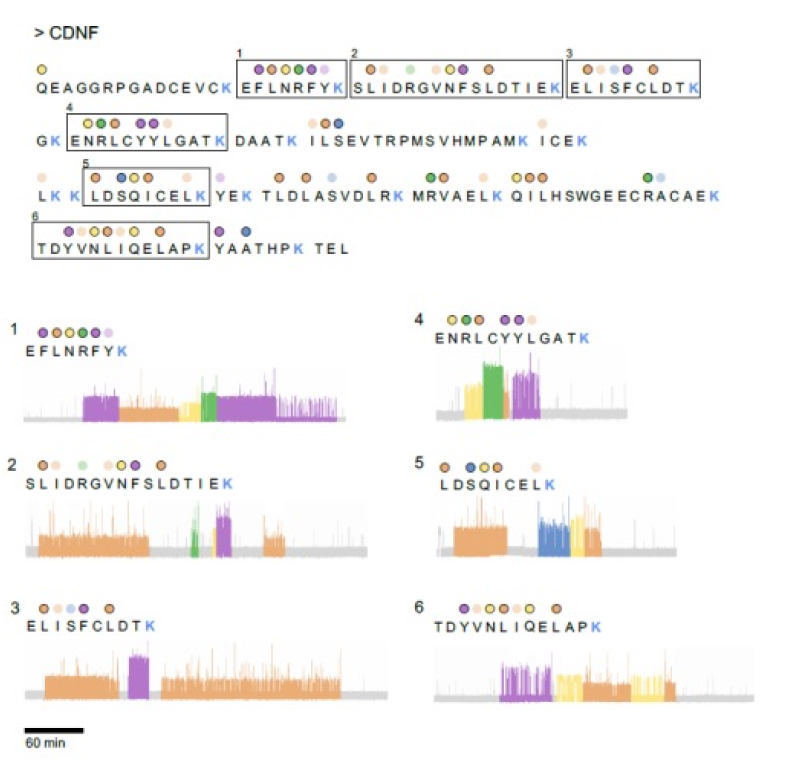
Here we showcase CDNF peptide identification results from mass spectrometry and Platinum sequencing. Platinum sequencing resulted in unique identification of 6 peptides and demonstrates Platinum’s ability for accurate protein identification through a simplified workflow.
-
Protein Identification using Next-Generation Protein Sequencing of In-Gel Digested Proteins – TECH NOTE
Western blot, a prevalent method for protein identification, often faces challenges due to the need for quality antibodies and the inability to differentiate similar-sized proteins. This application note introduces a non-antibody-based approach, detailing an in-gel protein digestion and peptide extraction procedure. This innovative method integrates with Quantum-Si’s library preparation and protein sequencing workflows, allowing for the separation, enrichment, and identification of proteins using Quantum-Si’s Platinum next-generation protein sequencing™ instrument.
Curious about revolutionizing your protein identification process? Download the full application note to dive deeper!
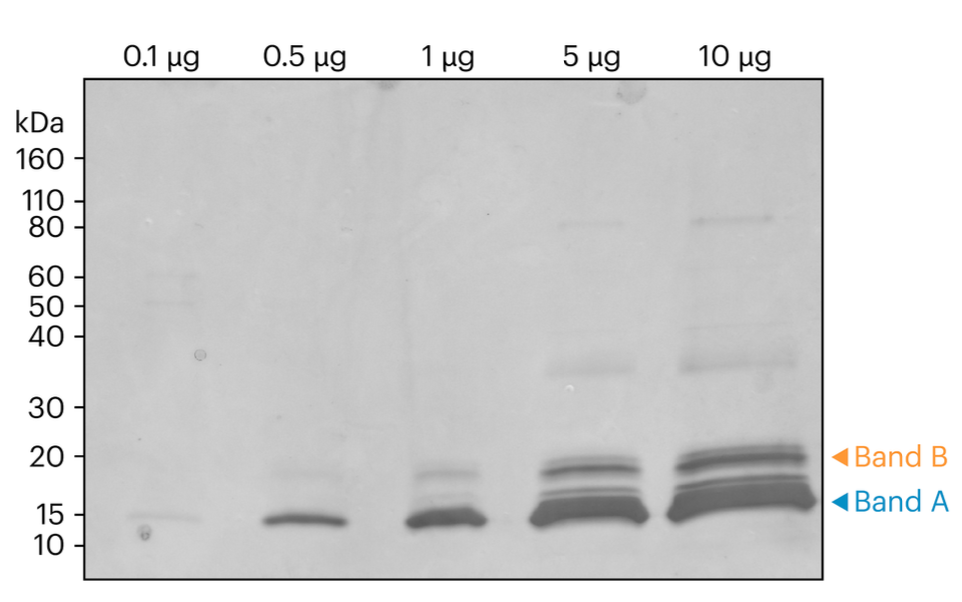
Figure 1. Here we showcase an SDS-PAGE representation of CDNF Samples before the band excision process. The visual provides a clear indication of the bands that were selected for in-gel digestion and subsequent sequencing, marked by distinct orange and blue arrows. This serves as a foundational step in understanding the sample preparation process.
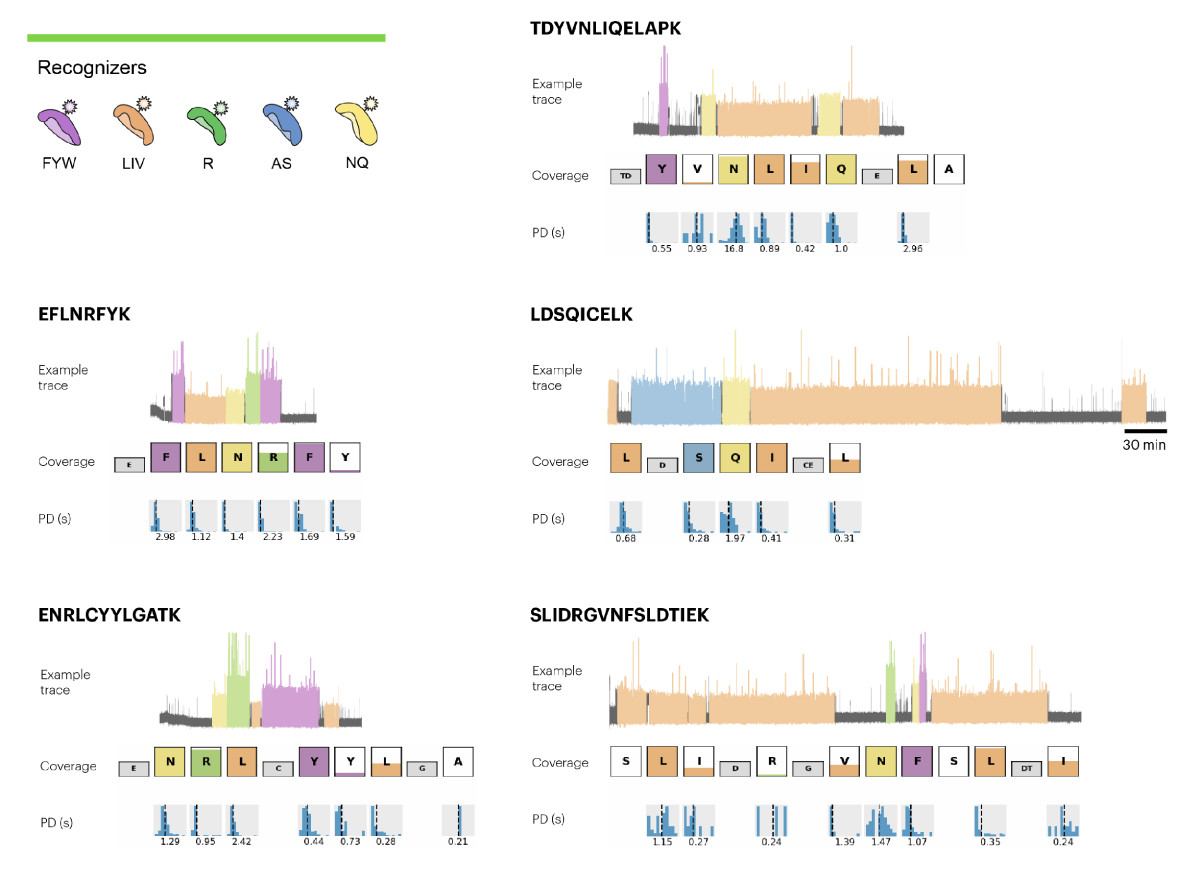
Figure 2. Representative Traces, Coverage, and Pulse Duration Data for the Peptides Identified in the 10 μg Sample of CDNF. Five recognizers were used to identify 12 amino acids (F, Y, W, L, I, V, A, S, N, Q, R, and K), enabling identification of 5 peptides as shown in the figure.
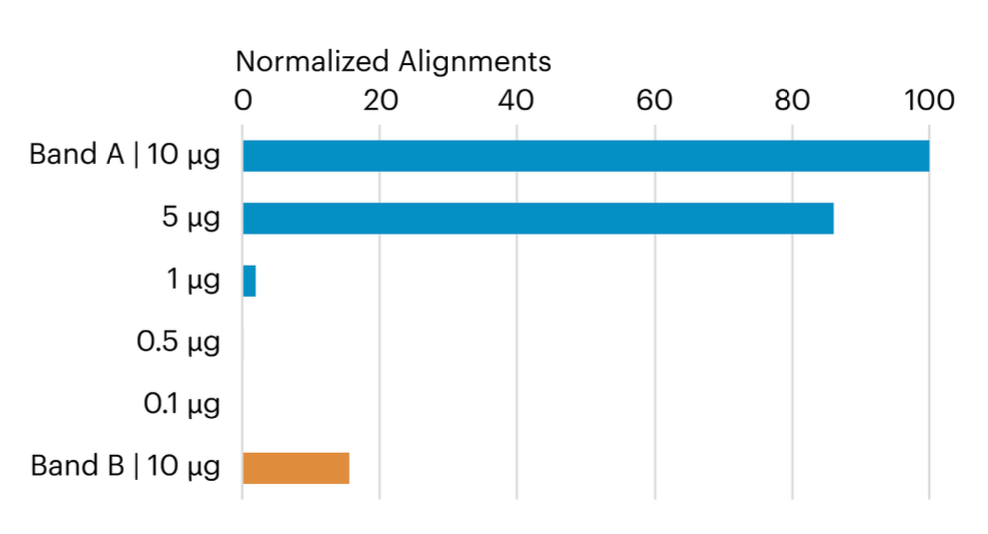
This graph compares the relative number of peptide alignments for each in-gel digested sample and reveals that both the 5 µg and 10 µg samples produced a higher number of aligned CDNF peptides compared to the smaller samples. Additionally, the 10 µg Band B sample also yielded a significant count of CDNF-aligned peptides, providing valuable insights into the efficiency and results of the in-gel digestion process.
-
Sequencing and Identification of Proteins from Five-Protein Mixtures on Platinum – APP NOTE
This application note introduces Quantum-Si’s Platinum instrument, which allows for the direct sequencing and identification of proteins in complex mixtures. By eliminating the need for targeted antibody-based assays, this technology offers a solution to detect multiple proteins simultaneously. We present a method for optimizing sample preparation and demonstrate the feasibility of identifying five distinct recombinant proteins in a mixture.
Download the full application note to learn more about this groundbreaking protein sequencing platform.
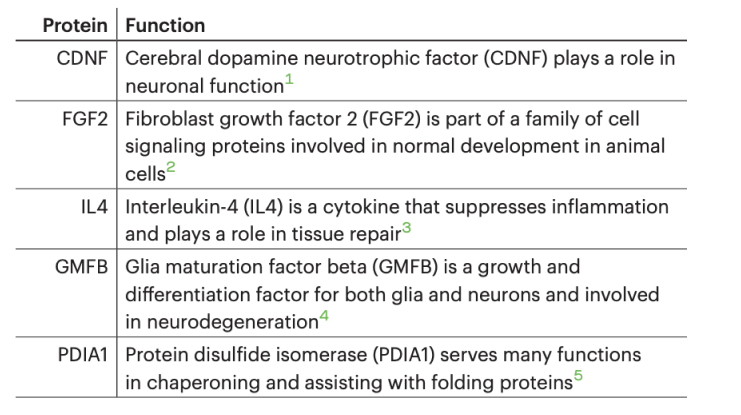
We tested the feasibility of detecting multiple proteins on Platinum by combining a mixture of five recombinant proteins (CDNF, FGF2, IL4, GMFB, and PDIA1) and employing protein sequencing to identify them.
To determine if overall peptide coverage would be affected by different library preparation methods, we compared two different mixes with increased complexity.
Read the application note for an in-depth discussion of our methods and results.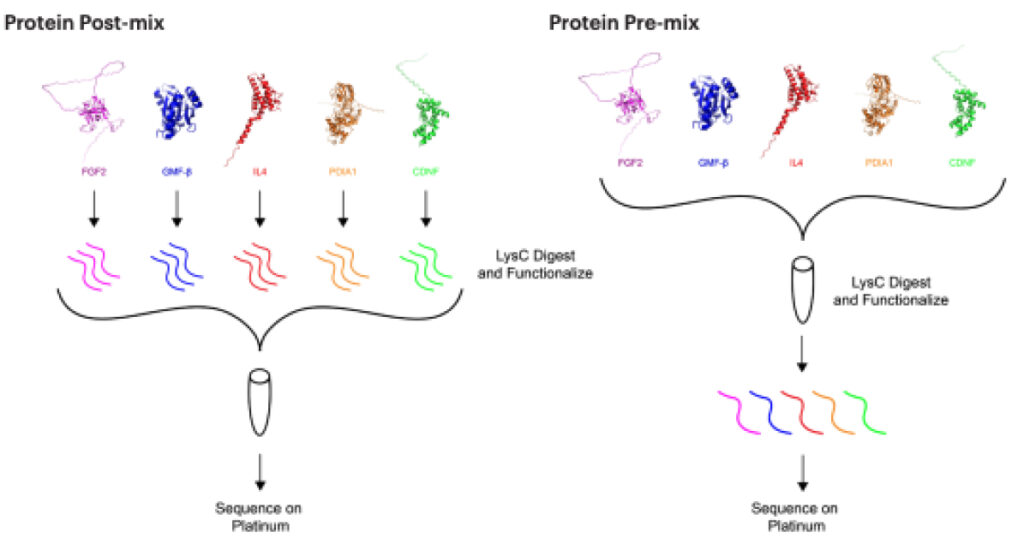
-
Characterization of Serum Proteins by Coupling Protein Separation Techniques with Next-Generation Protein Sequencing on Platinum — APP NOTE
Proteins play a vital role in virtually all biological processes, serving as key regulators of cellular functions, disease biomarkers, and therapeutic targets. However, the identification, isolation, and characterization of proteins from biological specimens can be challenging due to their complexity and diversity. Next-Generation Protein Sequencing (NGPS) on Platinum® offers a groundbreaking approach to deeply interrogate individual proteins with high resolution on a user-friendly benchtop platform. When combined with protein separation or enrichment methods, Platinum enables characterization of individual proteins from biological samples, revealing crucial details such as amino acid variants and PTMs. In this application note, we demonstrate a method that combines Sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) protein separation and NGPS on Platinum to identify and characterize serum proteins. This method holds potential for applications beyond serum, offering new insights into disease mechanisms and advancing therapeutic and diagnostic developments.
Download the full application note to learn more.
-
Identifying Monoclonal Antibodies with Quantum-Si’s Next-Generation Protein Sequencing™ Technology – APP NOTE
Identifying low-abundance monoclonal antibodies within a polyclonal antibody population is crucial for understanding disease mechanisms, diagnostics, and more. Traditional methods, while useful, often come with high costs, complexity, and time-consuming procedures. This application note introduces Quantum-Si’s next-generation protein sequencing™ technology on the Platinum instrument, a solution that addresses these challenges and efficiently identifies these critical antibodies.
Discover the breakthrough methodology and its potential impact by downloading the full application note.
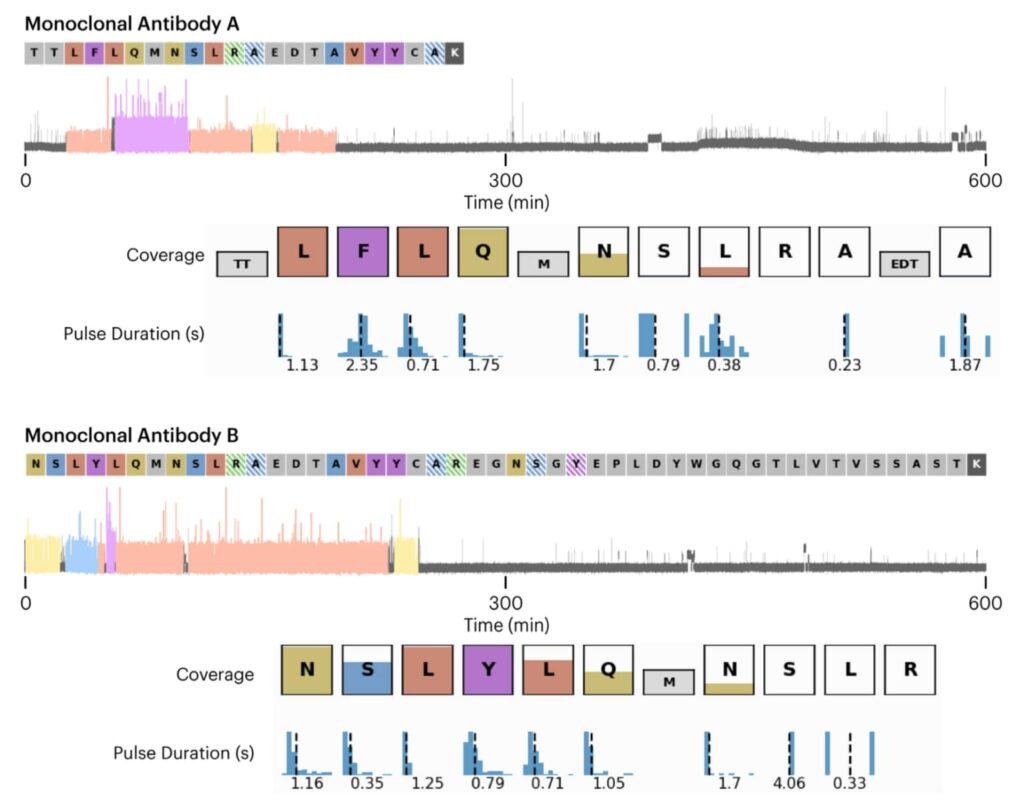
Here we showcase the sequencing data of unique peptides derived from two distinct monoclonal antibodies. The data, collected over a 10-hour period, displays the recognition segments (RS) with their coverage indicated by the color height in each box, alongside the pulse duration distribution. This visualization effectively demonstrates the Platinum instrument’s capability to accurately sequence and identify specific peptides unique to each antibody.
-
Next-Generation Protein Sequencing™ Platform: Advances in Protein Identification & Characterization — WEBINAR Dr Danielle Tullman-Ercek, Professor of Chemical and Biological Engineering, Northwestern University and Katherine Johnson, Senior Director, Product, QSI October 6, 2023
In this expert led webinar, Dr Danielle Tullman-Ercek, Professor of Chemical and Biological Engineering at the Northwestern University, and Katherine Johnson, Senior Director, Product, at Quantum-Si, Inc will discuss a novel next-generation protein sequencing™ platform, Platinum®, and how it can complement immunoassays and other proteomic workflows by providing deeper insights into proteins. Learn how this next-generation protein sequencing™ technology works and what applications can be performed using this novel technology. Additionally, Dr. Tullman-Ercek will illustrate how protein identification on Platinum was utilized in her research to provide additional information about proteins typically studied via conventional western blot and mass spectrometry techniques. She will demonstrate how next-generation protein sequencing™ provides additional information about proteins, comparing results from conventional techniques to those obtained from next-generation protein sequencing™.
Key learning objectives
- Learn about Next-Generation Protein Sequencing, how it works and key benefits
- Explore how it compares to and complements conventional proteomic techniques such as mass spectrometry and immunoassays
- Explore the applications that can be performed with Platinum next-generation sequencing technology
- Discover how Platinum is utilized to complement data obtained from conventional protein identification techniques to provide deeper insights into proteins
Speakers: Dr Danielle Tullman-Ercek, Professor of Chemical and Biological Engineering, Northwestern University and Katherine Johnson, Senior Director, Product, QSI Date: October 6, 2023
-
Quantum-Si’s Next-Generation Protein Sequencer™ Enables Protein Detection and Peptide Characterization — POSTER US HUPO | 2024
Sequencing proteins and correlating amino acid changes to biological function is critical to advancing our understanding of human health and disease. Next-Generation Protein Sequencing™ (NGPS) on Platinum® enables researchers to identify and characterize proteins with single-molecule resolution in a simple workflow and on a benchtop instrument. To demonstrate the versatility of Platinum and the use of Kinetic Signatures, we sequenced various types of samples, including recombinant proteins, protein mixtures, immunoprecipitated proteins, peptide barcodes, and peptides with Post Translational Modifications (PTMs).
Furthermore, we showcased the detection of peptide barcodes along with the utilization of barcoding techniques to streamline protein engineering applications.
Recent product enhancements to the platform include new sequencing kit and chip chemistry as well as advanced analytical software tools. These enhancements unlock new applications such as the use of protein inference to study unknown protein samples.